Set up a Docker run command to run your script on some data
Docker run command overview
The docker run command is used to create a container based on the specified container image. What the launched container does depends on the additions and options you send to the command.
The format of the docker run command is like this:
docker run [OPTIONS] imagetag:imageversion [COMMAND] [ARG...]
Where you would make the following replacements:
[OPTIONS]gets replaced with any number of option flags that thedocker runcommand can take inimagetag:imageversiongets replaced with the tag and version of an image you want to run a container of[COMMAND]gets replaced with a command you want to run within the container you launch (We will use this, but it is technically optional)[ARG...]gets replaced with any arguments to your command from[COMMAND](This is only needed if you are including a[COMMAND])
There are a number of options you can include with the docker run command, which you can investigate in the Docker run command documentation. For now we will go through some simple examples that are relevant to this tutorial.
Basic docker run command with our example
A very basic Docker run command with no options included, and using the tutorial image name and tag, looks like this:
docker run container_tutorial:1.0-dev
We’re specifying that we want docker run to create a container from our image container_tutorial:1.0-dev.
If you enter that in your command line and press Enter, this command will launch a container from our tutorial container image. However, it will stop immediately and finish the command pretty quickly:

since no input options or within-container [COMMAND] were specified. Some more specific options are required to work with a container you create.
The main examples we will look at are interacting with a container directly, and running a specific command in a container. To run your workflow script we will do the latter: send a specific command-line command to your container. First we’ll take a quick look at how to run a container in interactive mode to take actions directly within a running container.
Interacting with a container directly (Interactive mode)
Later in this tutorial we’ll be creating docker run commands that tell Docker Engine to create a container from your image and send a single command directly to that container. Here is a quick demo of another way to work with a container: you can run a container from your image in interactive mode.
The option flags -i and -t can be used with docker run and your container tag, like this:
docker run -i -t container_tutorial:1.0-dev
Running that command will start up a container based on the image tagged container_tutorial:1.0-dev and run it in your command line in interactive mode. When you enter that command and press Enter, you’ll see a command prompt with a different label that indicates you are inside a running container from your image.

You are inside an instance of your container! You now have an input/output terminal inside an instance of your container image, and all the instructions you entered in your Dockerfile have been incorporated into this environment.
Our Dockerfile example is built on top of the base container “freesurfer/freesurfer:7.3.2”, which uses CentOS as its operating system, and uses a bash shell by default. This means you can run bash shell commands for a CentOS Linux system from here, as long as the commands either come with the operating system by default or have been installed via instructions in your Dockerfile. You can get the default working directory1 that you have automatically been placed into with pwd:

You can also get operating system info with cat /etc/os-release:

If we had added an entrypoint or initial command to our container build Dockerfile with the ENTRYPOINT2 or CMD3 instructions, running our container would execute those commands by default (in slightly different ways depending on whether you used ENTRYPOINT or CMD). We didn’t add either of those instructions, so no commands will get run by default when you start an instance of your container - in this case, starting a container instance with your docker run command will just start a container from it with no commands sent to it.
Since there aren’t any commands to run or complete from the Dockerfile or specified in the docker run command - you just have the container open in interactive mode - the container will just keep running.
Check that out by opening another terminal on the same machine you are working on, and run the command:
docker ps
The output of this command will display the list of currently running containers on your system. You should see a single instance of your container image running.

When you run a container from an image, Docker gives the container a name4 that you can then use to terminate the container with the command
docker kill CONTAINER_NAME
In this case, my auto-generated container name is charming_margulis so I would terminate my endless container with
docker kill charming_margulis
If you return to the terminal window where you were in your container, you’ll be able to see that the container process terminated when you ran the docker kill command from the other window, and you’ve been returned to your standard command prompt on your local system.
Goodnight, charming_margulis.
Running a specific command within a container
Next we’ll be setting up docker run commands that create a container from your image and then run a single command in that container.
Here is a quick reminder of the format of the docker run command:
docker run [OPTIONS] imagetag:imageversion [COMMAND] [ARG...]
We want to send a command in the [COMMAND] section, and use our image tag. Adding options in the [OPTIONS] part is optional so we’ll leave it out for now, for simplicity.
Sending a command would be formatted like this:
docker run container_tutorial:1.0-dev [COMMAND]
The text [COMMAND] can be replaced with any command-line command you want to run within your container. It will run it exactly as though you were running it in interactive mode in the previous section. Eventually we’ll use your custom workflow script command in place of [COMMAND], but for now we will show some simpler examples.
Print the default working directory in the container with pwd
First we’ll send the same first command we tried in interactive mode: showing the default working directory in the container by sending in the pwd command. Using our example build tag from earlier, and the command pwd, our run command will look like this:
docker run container_tutorial:1.0-dev ls
This will run a container based on your container_tutorial:1.0-dev image, run that pwd command, and then end the container process. It will show you the output from the container, and then end the container process so you can run another command.

List files in the container with ls
You can list the files in that default working directory of your container by sending in the ls command like this:

That command lists the files as though you were located in the default directory in interactive mode. In this example, that directory contains a default Documents folder that was originally in the base container and also some leftover files from the installation steps that were in the Dockerfile.
We could have copied our custom workflow script into the default directory and it would have shown up in that file list. But in the tutorial Dockerfile example, the COPY instruction that copied our run_workflow.sh script file into the container did this instead:
COPY run_workflow.sh /usr/local/bin/run_workflow.sh
The run_workflow.sh file got copied into /usr/local/bin. So if we list the files in /usr/local/bin within our container, we should see that file name in the list. Here is the command to run an instance of our container image and then list the files in that specific directory:
docker run container_tutorial:1.0-dev ls /usr/local/bin
Here is what the output looks like:

Our script file run_workflow.sh shows up in that folder.
Show environment variables with env
Another useful bash shell command to run before we go further is to check out what environment variables are set within the container with env, like this:
docker run container_tutorial:1.0-dev env
This will show a list of all the environment variables that are currently set in the container environment, like this:

Some of these came from the original FreeSurfer freesurfer/freesurfer:7.3.2 base container and carried over when we built on top of it. Some of the FSL environment variables are ones that we set in these lines of the original Dockerfile:
ENV FSLDIR /usr/local/fsl
ENV FSLMULTIFILEQUIT TRUE
ENV FSLOUTPUTTYPE NIFTI_GZ
ENV PATH $FSLDIR/bin:$PATH
The FSL and FreeSurfer-specific environment variables will be important for running functions from those programs, and the PATH variable is important since that is a list of folders where our container will look for program files5, for example where it will look for programs we installed like fslmaths. In fact, you can 100% confirm that your container knows where fslmaths is by running
docker run container_tutorial:1.0-dev fslmaths
You’ll see the fslmaths help block:

Get your container to see your local data
The container is an isolated environment that is entirely separate from your local files, programs, environment variables, and data. This means that by default, your container has no access any files that haven’t been copied in via a Dockerfile RUN instruction. You’ll need to run your workflow script on specific imaging data, so you need a way to get your imaging dataset into the container.
You could technically copy it all in with a bunch of Dockerfile RUN commands during your docker build step, but that’s not a great idea - it’ll increase the container image size considerably, plus you want to be able to share your container image but not give the world access to your dataset. A relatively simple way to get your container to see your local files without permanently storing them in the container image is to “mount” a folder with your data in it into your container with a “bind mount”.
The -v flag is the option flag that can be used to mount data into your container. When mounting a folder with -v, you send the -v option as part of the docker run command options section like this:
docker run -v A:B container_tutorial:1.0-dev [COMMAND]
To mount a folder, -v should be followed by a string in the format A:B as shown above. A needs to be replaced with the absolute path (starting with a /) to the folder on your local computer that you want your Docker container to see. B should be replaced with the absolute path (starting with /) of where you want that folder to “appear” within a running container (so the location where your container will think that folder is).
Here is that sample command with -v shown with a more accurate-looking example of the A and B folder paths
docker run -v /full/path/to/localfolder:/containerdata container_tutorial:1.0-dev [COMMAND]
In this sample command, any files that are located on your computer at the path /full/path/to/localfolder will show up in the folder /containerdata in Docker.
Here is what it looks like with real data. Use cd to get into your main containerization_tutorial directory. This is the one that contains subfolders for scans, freesurfers, container_image_files, and output, as well as the example scripts.

For your script you’ll need the folders with your example data, plus you’ll want to send an output folder for script output to go into. So ultimately you want to mount the freesurfers, scans, and output folders into your container. On your local computer, these folders are subfolders your current working directory containerization_tutorial. Use pwd to get the absolute path of your containerization_tutorial directory.
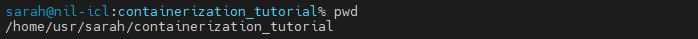
This is the path you’ll want to use as the basis for the A path - the path to your local folder with your data on your computer. In this case, my containerization tutorial directory is at /home/usr/sarah/containerization_tutorial. My data is in these subfolders:
- Scan files are in
/home/usr/sarah/containerization_tutorial/scans - FreeSurfer files are in
/home/usr/sarah/containerization_tutorial/freesurfers - The empty output directory I created is in
/home/usr/sarah/containerization_tutorial/output
Now for choosing the B path. This can be any folder path that doesn’t already exist within the container. It can be any path because you’ll be telling your script to look there anyway, since a couple of your script input parameters are paths to a folder of scans, path to a folder of freesurfers, and path to an empty output directory. It has to be an absolute path starting with /. For mapping the scans directory I’ll map it to the very simple path /scans - A folder called scans located in the base / directory in the container folder structure.
Here is a docker run command that will mount my example local scans folder into the container at path /scans, and then run the ls / command within that container:
docker run -v /home/usr/sarah/containerization_tutorial/scans:/scans containerization_tutorial:1.0-dev ls /
In this command, ls / will list the files in the / folder within a running container, and then the container will stop. In the output of the list, you’ll see the folders Docker can see - including scans folder you mounted with -v!

You can ls the /scans folder within the container to show what Docker sees in the /scans folder with
docker run -v /home/usr/sarah/containerization_tutorial/scans:/scans containerization_tutorial:1.0-dev ls /scans
You’ll see the session folder:

And when the container finishes running that command, if you list the scans that you can see locally, you’ll see the same thing Docker sees.

with that -v command and the correct absolute paths, Docker can now see the contents of your containerization_tutorial/scans folder.
To map multiple folders in a docker run command, you add extra -v A:B options like this:
docker run -v A1:B1 -v A2:B2 -v A3:B3 container_tutorial:1.0-dev [COMMAND]
Here is my example with the scans folder mapped to /scans in the container, freesurfers mapped to /freesurfers, and output mapped to /output:
docker run -v /home/usr/sarah/containerization_tutorial/scans:/scans -v /home/usr/sarah/containerization_tutorial/freesurfers:/freesurfers -v /home/usr/sarah/containerization_tutorial/output:/output container_tutorial:1.0-dev ls /
The ls / will show the list of folders the container can see and include the newly mounted freesurfers and output directories you added on.
Setting up a command to run your script within the container
Now that you can send your files to the container it’s time to set up a run command specific to your workflow script. As you can see the run command has a bunch of different parts and can get pretty long, so it’s useful to set up your command in a text editor and then paste it into your command line when you are sure it’s ready to run.
You’ll want to combine the docker run command with the correct mounted data folders with the correct [COMMAND] to run your workflow script within the container.
Here is a reminder of how to call your run_workflow.sh script after the last part of the the workflow script setup:
./run_workflow.sh scans_dir freesurfers_dir output_dir session_label dti_scan_id dti_scan_filename dti_bvec_filename dti_bval_filename fi_threshold_value gradient_value
Here is the script command with your specific inputs that you tested earlier from your containerization_tutorial folder:
./run_workflow.sh scans freesurfers output OAS30001_MR_d3132 dwi1 sub-OAS30001_sess-d3132_run-01_dwi.nii.gz sub-OAS30001_sess-d3132_run-01_dwi.bvec sub-OAS30001_sess-d3132_run-01_dwi.bval 0.25 0
Now to adapt this to run within the container.
As mentioned in an earlier section, your Docker file copied the run_workflow.sh into /usr/local/bin. You can confirm again with this container run and list command:
docker run container_tutorial:1.0-dev ls /usr/local/bin
So when you call your script command in the container, you’ll use /usr/local/bin/run_workflow.sh instead of ./run_workflow.sh, like this:
/usr/local/bin/run_workflow.sh scans_dir freesurfers_dir output_dir session_label dti_scan_id dti_scan_filename dti_bvec_filename dti_bval_filename fi_threshold_value gradient_value
The first 3 inputs to your script command are the scans_dir for scan file directory, freesurfers_dir for the freesurfers directory, and output_dir for the directory to send output files to. Now you know how to mount those three folders into a running instance of your container. When you update your script call for your in-container run, you’ll enter Docker’s mounted locations of those folders as the inputs. So since Docker sees your local scans folder at /scans, your local freesurfers folder at /freesurfers, and your local output folder at /output, you would send the folder paths Docker can see as those inputs to the script, like this:
/usr/local/bin/run_workflow.sh /scans /freesurfers /output session_label dti_scan_id dti_scan_filename dti_bvec_filename dti_bval_filename fi_threshold_value gradient_value
That takes care of where the inputs and outputs come from - every other input to your script can be entered the exact same way as if you were running locally:
/usr/local/bin/run_workflow.sh /scans /freesurfers /output OAS30001_MR_d3132 dwi1 sub-OAS30001_sess-d3132_run-01_dwi.nii.gz sub-OAS30001_sess-d3132_run-01_dwi.bvec sub-OAS30001_sess-d3132_run-01_dwi.bval 0.25 0
That’s the container-specific [COMMAND] part you want to send to your docker run command. Combine that with the command that mounts your data directories to get the full run command for your custom workflow:
docker run -v /home/usr/sarah/containerization_tutorial/scans:/scans -v /home/usr/sarah/containerization_tutorial/freesurfers:/freesurfers -v /home/usr/sarah/container_tutorial/output:/output container_tutorial:1.0-dev /usr/local/bin/run_workflow.sh /scans /freesurfers /output OAS30001_MR_d3132 dwi1 sub-OAS30001_sess-d3132_run-01_dwi.nii.gz sub-OAS30001_sess-d3132_run-01_dwi.bvec sub-OAS30001_sess-d3132_run-01_dwi.bval 0.25 0
In each of the -v inputs, you’ll need to replace /home/usr/sarah/ with the absolute path to where your containerization_tutorial directory is located, to tell Docker to mount your data locations on your specific system.
Run your run command
Since the -v mount option uses absolute paths, you can actually run this docker run command from any folder location on your machine. Paste your entire customized docker run command into the command line and press Enter. You should see output from the commands on your screen:

And when it’s done, you should see the expected output files appear in your local output folder:

-
You can actually see where the FreeSurfer team set the default directory by looking at the Image Layers for the freesurfer/freesurfer:732 image on Docker Hub.. ↩
-
More details on Docker ENTRYPOINT setup within a Dockerfile can be found in the Dockerfile reference guide.. ↩
-
More details on Docker CMD setup within a Dockerfile can be found in the Dockerfile reference guide.. ↩
-
A quick overview of the PATH variable can be found on Wikipedia and in many other Unix resources elsewhere.. ↩